Model assessment of reductive dechlorination as a remediation technology for contaminant sources in fractured clay: Modeling tool
Appendix E Sensitivity analysis
The sensitivity analysis is applied to a “base case”, and the change in the output concentrations, due to change in one parameter, is calculated. The “base case” parameters are chosen, in order to obtain output curves, close to the one obtained with experiments. The “base case” parameters are shown in Table E.1. It has to be noticed that a same specific yield value is assumed for both biomass groups. Furthermore, X10 and X20 are the initial biomass concentrations. While no prior information is available on the initial concentration of group 1, the initial concentration of group 2 can be estimated. Hence this group represents the concentration of Dehalococcoides, which has been added to the sample. Based on the estimated amount of Dehalococcoides cells in the KB-1 culture, and the dilution factor, it is possible to have an estimation for X20. However, this number is subject to uncertainty, that is why it is considered as a parameter to optimize.
The base case is based on the experimental protocol followed during the treatability study. This means that the 2nd biomass group is assumed to be introduced first 57 days after the beginning of the experiment. Furthermore, the initial TCE concentration is set to 15 µM, as this value is close to the ones observed in experiments.
E.1 Estimation of X2o
A cell density of 108 cells/mL is commonly used for KB-1 culture [Dennis, pers. communation, 2008], of which a varying amount is Dehalococooides. During microcosm experiments, the culture is commonly diluted between 300 and 3000 times [Jørgensen et al., 2005], resulting in an initial concentration in Dehalococcoides between 107 – 4*108 cells/L.
Table E.1 - Base case parameters
| Units | Base case | |
| µTCE | d-1 | 2 |
| µDCE | d-1 | 0.1 |
| µVC | d-1 | 0.1 |
| KTCE | µmol.L-1 | 10 |
| KDCE | µmol.L-1 | 3.3 |
| KVC | µmol.L-1 | 2.6 |
| Ki,TCE | µmol.L-1 | 10 |
| Ki,DCE | µmol.L-1 | 3.6 |
| Ki,VC | µmol.L-1 | 7.8 |
| Y | cell.µmol-1 | 5.2*108 |
| kd1 | d-1 | 0.03 |
| kd2 | d-1 | 0.03 |
| X10 | cell.L-1 | 8*104 |
| X20 | cell.L-1 | 2.5*108 |
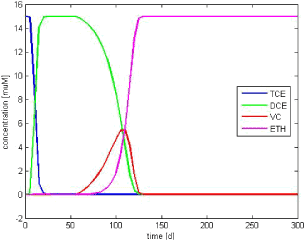
Figure E.1 – Base case simulation results
E.2 Sensitivity assessment
The sensitivity is assessed by calculating the sensitivity index S, related to the variation of each parameter [Spitz and Moreno, 1996]:
![]()
Where |dCi|is the difference in concentration of compound i between the base case and sensitivity case, dP is the change in input parameter and P is the initial input parameter value.
E.2.1 Parameter value range in literature
In Table E.2, the ranges found in the literature for the different parameters are shown. The ranges for some of the parameters are very wide, with several orders of magnitude. It has to be noticed that these parameters are reported for different cultures, electron donors and experimental conditions.
Table E.2 - Range of parameters to optimized in literature
| Units | Range in literature | |
| µTCE | d-1 | 0.013 – 4.3 |
| µDCE | d-1 | 0.003 – 0.766 |
| µVC | d-1 | 0.003 – 0.737 |
| KTCE | µmol.L-1 | 0.05 – 17.4 |
| KDCE | µmol.L-1 | 0.54 – 11.9 |
| KVC | µmol.L-1 | 2.2 – 602 |
| Ki,TCE | µmol.L-1 | 0.05 – 724 |
| Ki,DCE | µmol.L-1 | 1.8 – 600 |
| Ki,VC | µmol.L-1 | 2.6 – 602 |
| Y | cell.µmol-1 | 4.3*108 – 1.9*109 |
| kd1 | d-1 | 0.01 – 0.05 |
| kd2 | d-1 | 0.01 – 0.05 |
| X10 | cell.L-1 | - |
| X20 | cell.L-1 | - |
As it is more likely that the parameters will remain in the range of the base case values, two different sensitivity values are computed: one for parameter change within the literature range (max and min values) and the other for changes of +/- 50% from the base case values.
E.2.2 Sensitivity analysis results
The sensitivity indexes range between 0 and 250, there are some differences between the +/- 50% scenario and the max/min literature values scenario. These differences come from the wide range in which some parameters vary in the literature, for example µDCE and µVC varies by more than 3 order of magnitude (between 0.003 to 0.8 d-1) However, the seven most sensitive parameters are the same, i.e. µDCE, Y, X20, kd2, µVC, KDCE and µTCE. The optimization should then focus on these seven parameters to achieve relevant results.
Table E.3 - Normalized sensitivity index for +/- 50% variation and literature extreme values
| +/- 50% | Max/min in literature | |
| µDCE | 161 | 163 |
| Y | 37 | 26 |
| X20 | 37 | 61 |
| kd2 | 34 | 35 |
| µVC | 32 | 143 |
| KDCE | 27 | 28 |
| µTCE | 24 | 226 |
| KVC | 17 | 10 |
| Ki,DCE | 16 | 12 |
| KTCE | 10 | 8 |
| Ki,VC | 8 | 8 |
| X10 | 5 | 2 |
| kd1 | 2 | 2 |
| Ki,TCE | 0 | 0 |
E.3 Experimental data from [Friis et al., 2007]
In these experiments, electron donor and KB-1 culture are added at the beginning of the experiments. Furthermore, there are no bacteria present in the bottle prior to the addition of KB-1. Hence, it is possible to estimate the different biomass populations, from the estimated cell density of KB-1 culture. In these experiments, the culture is diluted 300 times [Friis et al., 2007], leading to an initial Dehalococcoides concentration (X20) between 108 and 109 cells/L, and other biomass population (X10) approximately 2.3*108 to 2.3*109 cell/L.
Among the seven parameters to optimize, it can be assumed that only the three maximum growth rates are different from one experiment set to the other (with different electron donor and/or temperature). Hence, it is expected that the four other parameters have a common optimized values, valid for all the experiments.
The optimization script aims at reducing the root mean squared error (RMSE), as defined by:
![]()
As explained previously, lactate is a fast hydrogen release, while propionate releases hydrogen much slower. Hence, it is expected that the experiments with lactate-amended culture correspond better to the no-limiting substrate assumption. Therefore it was decided to optimize all parameters with lactate-amended results and then to use the optimized values of KDCE, Y, X20 and kd2 for results with propionate, and optimize only the growth rates.
Version 1.0 July 2009, © Danish Environmental Protection Agency