[Front page] [Contents] [Previous] [Next] |
Vibrio vulnificus in Denmark
2. Identification of Vibrio vulnificus
2.1 Classification and taxonomy
V. vulnificus is a halophilic marine bacterium (Baumann & Schubert, 1984). As other members of the genus Vibrio, family Vibrionaceae, it is a Gram-negative rod, aerobic and facultatively anaerobic, motile by means of a polar sheated flagellum, and is oxidase and catalase positive (Baumann & Schubert, 1984).
The taxonomy of V. vulnificus was first investigated by Baumann et al. (1973). V. vulnificus was described as a group of Gram-negative, fermentative marine organisms which were assigned to group C-2. Hollis et al. (1976) designated the same organism as Alactose-positive Vibrio@ or AL+Vibrio@ since the ability to ferment lactose was one characteristic that could distinguish this species from Vibrio parahaemolyticus and Vibrio alginolyticus. Today it is known that the lactose fermentation reaction is negative in 25% of the V. vulnificus isolates (Baumann et al., 1971). Strains within group C-2 were found to be genetically related, based on DNA/DNA hybridization and were assigned as a new species designated Beneckea vulnifica (Avulnifica@ means wound in Latin) (Reichelt et al., 1976). Similar studies performed on the L+Vibrio concluded that this group was a species separate from V. parahaemolyticus and V. alginolyticus (Clark & Steigerwalt, 1977). In 1979, the transfer of Beneckea vulnifica (synonym = L+Vibrio) to the genus Vibrio was proposed and its name became V. vulnificus (Farmer, 1979). The name V. vulnificus was given official taxonomic status in 1980 (Farmer, 1980). It is believed that V. vulnificus formerly often was misidentified as V. parahaemolyticus (Hollis et al., 1976) The species V. vulnificus comprises two biotypes which in the original definition differed phenotypically, serologically, and in host range (Tison et al., 1982) (Table 1).
V. vulnificus biotype 1 is ubiquitous to estuarine environments and is an opportunistic human pathogen (Oliver, 1989). Biotype 2 is typically recovered from diseased eels but is also reported to cause illness in humans after handling eels (Mertens et al., 1979; Dalsgaard et al., 1996b). The division into biotypes is no longer supported and a division into serovars has been suggested (Høi et al., 1998b). The term Abiotype A is still used in the literature and will therefore also be used in this thesis. Chapter 5 includes the latest suggestions in this area and explains why Aserovar@ is a better term than Abiotype@.
Several studies have dealt with the phylogenetic relatedness of V. vulnificus and other Vibrio species based on sequencing of 5S, 16S and 23S rRNA as well as immunological relationship among superoxide dismutases (Baumann et al., 1980; MacDonell & Colwell, 1985; Dorsch et al., 1992; Aznar et al., 1994). Three types of cluster analysis based on 5S rRNA sequences all generated dendrograms where V. vulnificus showed the highest similarity to V. cholerae (MacDonell & Colwell, 1985). In another study V. vulnificus was described to be more closely related to V. parahaemolyticus based on 16S rRNA sequences obtained by reverse transcription of cDNA (Fig. 1) (Dorsch et al., 1992). A later more extensive study using alignment of 16S rRNA sequences reported that V. vulnificus did not belong to the core organisms of the genus Vibrio ( Fig. 2) (Aznar et al., 1994).
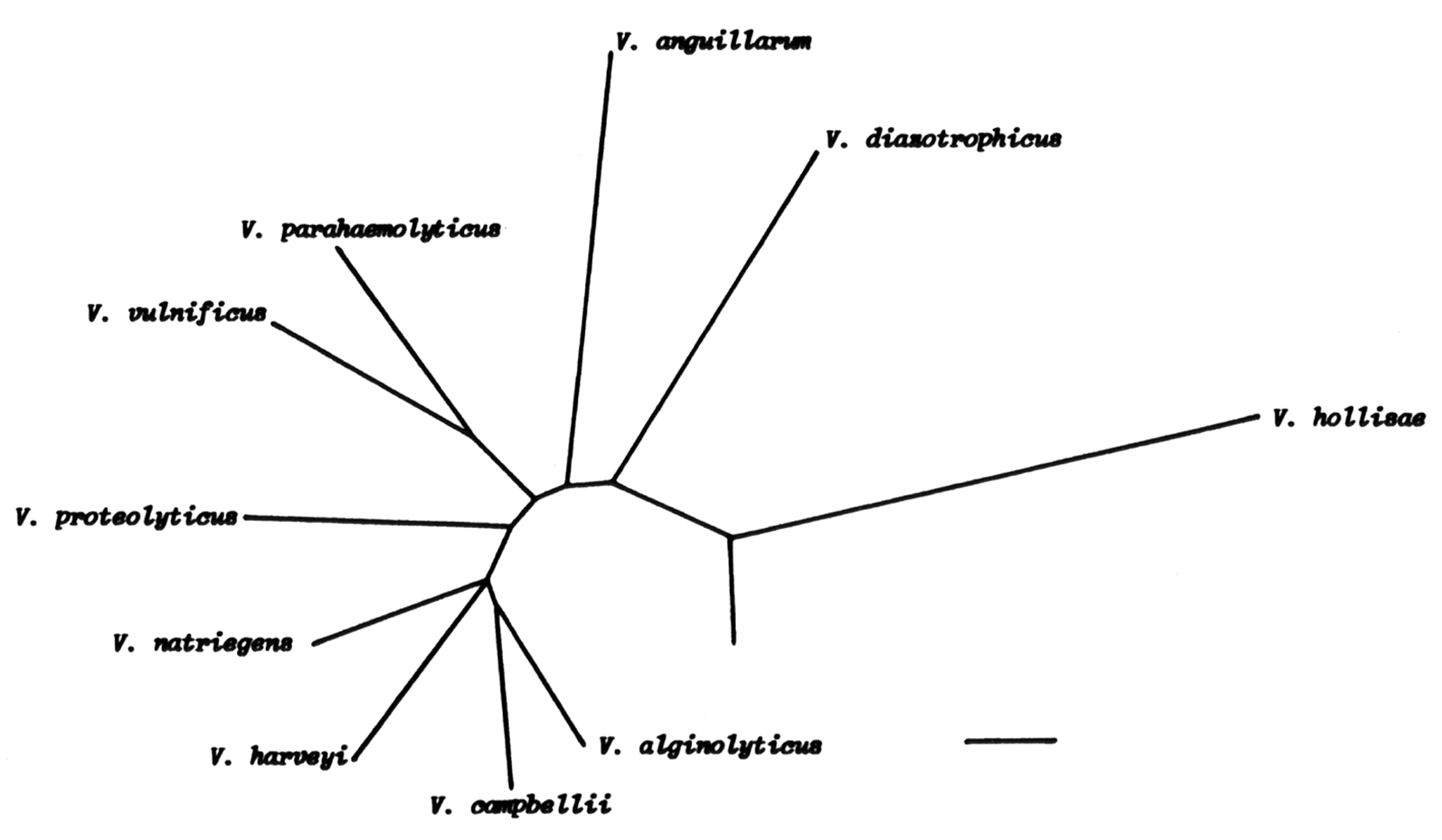
Figure 1: Phylogenetic relationship among 10 Vibrio species based on 16S rRNA sequences (Dorsch et. al., 1992). Bar = 10 nucleotide exchanges.
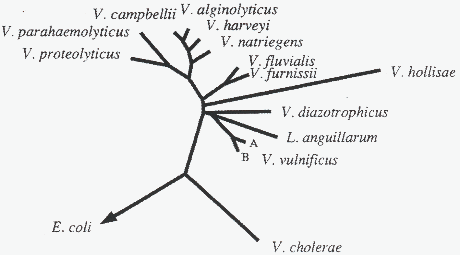
Figure 2: Phylogenetic tree showing the phylogenetic relationship of V. vulnificus and other Vibrio species based on 16S rRNA sequences. A and B represent two subgroups of 16S rRNA detected in V. vulnificus (Aznar et. al., 1994)
Data from comparative immunological studies of superoxide dismutases of vibrios supported that V. vulnificus is not closely related to the core organisms of the genus Vibrio (Baumann et al., 1980). Fig. 2 shows that two subgroups (A and B) among V. vulnificus were identified based on 16S rRNA alignments (Aznar et al., 1994). Strains of both biotypes belonged to group A and Aznar et al. (1994) concluded that the separation of V. vulnificus into two biotypes defined by biochemical and immunological properties does not reflect the genetical relationship of the strains. The phylogeny of the family Vibrionaceae, including V. vulnificus is at present not well established.
Table 1. Characteristics of V. vulnificus biotype 1 and 2 strains as originally defined by Tison et al. (1982). Values are percentage positive strains.
| Test |
V. vulnificus biotype 1 (9 strains) |
V. vulnificus biotype 2 (3 strains) |
| Indole production (Kovács) | 100 |
0 |
| Ornithine decarboxylase (Møller) | 89 |
0 |
| D-Mannitol | 44 |
0 |
| D-Sorbitol | 44 |
0 |
| Growth at 42°C | 44 |
0 |
| Pathogenicity for eels | 0 |
100 |
| Pathogenicity for mice | 100 |
100 |
| Agglutination with rabbit antiserum raised against biotype 2 strains | 0 |
100 |
2.2 Biochemical identification
Biochemical identification of V. vulnificus requires a number of biochemical assays which are costly and the result can not be obtained within the same day (Colwell, 1984; Murray, 1995). Table 2 shows selected biochemical characters of V. vulnificus and illustrates the difficulties in making a definitive identification based on biochemical reactions. Various references do not agree on reactions in certain biochemical tests. The API 20E index, which is only based on 31 V. vulnificus strains, differs from the reactions shown in three manuals in the lysine decarboxylase and Voges-Proskauer reactions (Table 2) (Barrow & Feltham, 1993;. Holt et al., 1994; Murray, 1995; Sinding, 1998). In general, reactions listed in reference manuals are based on a limited number of clinical strains and do not include the more heterogenous reaction patterns of environmental strains, e.g. 3% of 66 environmental V. vulnificus isolates were positive in arginine decarboxylase even though this is not described in Manual of Clinical Microbiology or the API 20E index (Table 2) (Dalsgaard et al., 1996a). Commercial kits for identification of Enterobacteriaceae can give false reactions when used to identify V. vulnificus since they may contain inadequate amounts of NaCl for growth of this halophilic Vibrio species (Horre et al., 1996). To avoid this problem, a minimum of 0.5 % NaCl must be present or added to the medium (McLaughlin, 1995). MacDonell et al. (1982) recommended an addition of 2% marine salts to the API 20E system and incubation at 22°C. False positive reactions may be produced when the bacterial concentration in the diluent broth is too high (Davies et al., 1995).
Suspected V. vulnificus isolates from environmental samples were more frequently identified as V. vulnificus by a DNA probe directed against the cytolysin gene than by the API 20E system (Dalsgaard et al., 1996a). Sixty-six isolates identified as V. vulnificus with the specific DNA probe were tested with the API 20E strip according to the manufacturers instructions (Table 2). A total of 29 API 20E profiles were obtained. Only four of these profiles, representing 20 isolates, reached the identification threshold of V. vulnificus. The reasons for these difficulties could be discrepancies between the species-specific reaction pattern in the API 20E database and the reactions given in standard identification tables, the heterogeneity of environmental strains, and the limited number of V. vulnificus strains (31 strains) included in the API 20E database (Dalsgaard et al., 1996a; Sinding, 1998).
Table 2. Biochemical characters of V. vulnificus (Barrow & Feltham, 1993; Holt et al., 1994; bioMérieux, 1995; Murray, 1995; Dalsgaard et al., 1996a)
Biosca et al. (1993a) reported that the API 20E system gave several false negative reactions when compared to conventional biochemically tests including the citrate test. Dalsgaard et al. (1996a) also reported that none of the 66 strains tested could utilize citrate in the API 20E test strip (Table 2). Changing the diluent in the API 20E from containing 0.9% NaCl (as recommended by the manufacturer) to 2% marine salts can promote citrate utilization (MacDonell et al., 1982).
Biotype 2 isolates are not included in the API 20E system and can not be identified by this system (Biosca et al., 1993a). The three manuals referred in Table 2 do not recognize the existence of V. vulnificus biotype 2 strains since they do not include indole negative strains.
Two studies have suggested that the API 20E can be used to identify the more common members of Vibrionaceae including V. vulnificus (Overman et al., 1985; Overman & Overley, 1986). These studies are based on a limited number of Vibrio sp. including one V. vulnificus strain each of unknown origin.
2.3 Identification using serological and molecular methods
The US Food and Drug Administration (USFDA) is currently using an enzyme immunoassay (EIA) in an ELISA format to identify presumptive V. vulnificus subcultured from modified cellobiose polymyxin B colistin (mCPC) agar (US Food and Drug Administration, 1995). The assay uses a V. vulnificus-specific monoclonal antibody (MAb) directed against an intracellular epitope of V. vulnificus (Tamplin et al.,1991; US Food and Drug Administration, 1995). No cross reactions to other Vibrio species and non-Vibrio species have been described and the ELISA format reduces assay time and facilitates handling of large numbers of test samples (Tamplin et al.,1991). The cell line producing the V. vulnificus-specific monoclonal antibody (MAb) is available at the American Type Culture Collection (ATCC).
Detection by ELISA method
Parker & Lewis (1995) developed a sandwich ELISA assay for detection of V. vulnificus in environmental specimens using anti-hemolysin as capture and detector antibody reagents. The authors claimed that the sandwich ELISA offers time-saving and labor-saving advantages over the currently accepted EIA but so far no other researchers have chosen to use the sandwich ELISA (Parker & Lewis, 1995).
Agglunitiation with anti-flagellar antibody
V. vulnificus can be identified one step beyond primary isolation with anti-flagellar (anti-H) antibodies (Simonson & Siebeling, 1986). The anti-H antibodies are produced in rabbits immunized with the flagellar core protein prepared from V. vulnificus. The antibodies are coated onto Staphylococcus aureus Cowan 1 cells to permit visual coagglutination within the time frame of the slide test. The agglutination reaction is fast and reliable and has been optimized to include the use of MAbs. The anti-core H antibodies are not available commercially so the technique includes purification of the flagellar protein and immunization of rabbits which is time-consuming (Simonson & Siebeling, 1986).
Detection with DNA probes
Molecular techniques, particularly specific oligonucleotide probes, constitute a very sensitive and specific tool for detecting V. vulnificus. An alkaline phosphatase-labeled oligonucleotide probe directed towards the cytolysin gene of V. vulnificus was constructed by Wright et al. (1985,1993). This probe, termed VVAP, demonstrated 100% specificity and sensitivity for clinical and environmental isolates of V. vulnificus and numerous investigators have shown that cytolysin is produced by all V. vulnificus strains, including both biotypes, and is species-specific (Kaysner et al., 1987; Morris et al., 1987a; Parker & Lewis, 1995; Biosca et al., 1996c). The sequence of the cytolysin gene has also been used for constructing primers for PCR identification (Brauns et al., 1991; Coleman et al., 1996). Two studies have argued that the use of the cytolysin gene as a target region in PCR amplification or as a target for an oligonucleotide probe is not suitable for the detection of V. vulnificus (Aznar et al., 1994; Arias et al., 1995). A non-essential gene, such as the cytolysin gene, could theoretically be lost or rearranged without affecting the viability of the bacteria. Instead, it has been suggested to use primers/probes directed against rRNA genes, since rRNA molecules are essential constituents of all living organisms and are present in growing cells in very high numbers (Aznar et al., 1994; Arias et al., 1995). A recent comparative study of the VVAP probe to an oligonucleotide probe directed against a sequence in the 16S rRNA region showed that the rRNA probe was slightly more specific and sensitive than the VVAP probe for identification of V. vulnificus (Biosca et al., submitted). The VVAP probe failed to detect two of 308 V. vulnificus strains tested and further hybridized with two isolates of 104 non-V. vulnificus strains tested (Biosca et al., submitted).
Colony hybridization
Identification of V. vulnificus with colony hybridization offers at least three advantages compared to biochemical testing: (i) V. vulnificus usually produces strong signals with virtually no background with alkaline phosphatase-labeled oligonucleotide probes facilitating interpretations of hybridization membranes, whereas biochemical reaction patterns can be difficult to interpret, (ii) identification of suspect isolates with oligonucleotide probes is more reliable than conventional biochemical identification (Dalsgaard et al., 1996a), (iii) colony hybridization is less time-consuming and cheaper than conventional biochemical identification, especially if the probes are used in a low concentration and reused as recommended by Høi et. al. (1998c).
VBNC state
During the colder months V. vulnificus apparently enters a viable-but-nonculturable (VBNC) state or at least a state where the bacterium cannot be cultured by ordinary bacteriological methods (Oliver, 1995). Detection by PCR or direct detection with flourescent tagged antibodies may solve the problem of nonculturability. However, the importance of VBNC cells in the ecology and virulence of V. vulnificus is not finally established and will be further discussed in section 3.1 (Brauns et al., 1991).
2.4 Conclusions from Chapter 2
Identification of V. vulnificus with the commercial biochemical testing kit API 20E is not reliable because V. vulnificus strains produce heterogenous biochemical reaction patterns. Serological identification requires preparation of antibodies since no commercial V. vulnificus-specific antibodies are available. Colony hybridization with a V. vulnificus-specific oligonucleotide probe is specific, fast and cost-effective. Identification of V. vulnificus with oligonucleotide probes or PCR primers directed against rRNA sequences is recommended.
[Front page] [Contents] [Previous] [Next] [Top] |