Occurrence and fate of antibiotic resistant bacteria in sewage
3. Effects of hospital and pharmaceutical waste effluent on the prevalence of resistant Acinetobacter in the recipient sewers
| 3.1 | Effects of hospital waste effluent |
| 3.2 | Effects of pharmaceutical waste effluent |
| 3.3 | Conclusions |
Waste effluent from hospitals contains high numbers of resistant bacteria 48,58,59 and antibiotic residues at concentrations able to inhibit the growth of susceptible bacteria 35. Accordingly, hospital waste effluent could increase the numbers of resistant bacteria in the recipient sewers by both mechanisms of introduction and selection for resistant bacteria. The effects caused by the introduction of hospital waste effluent into the sewage system were prior to our investigation. Furthermore, no comparison was made with the effects produced by waste effluent from other potential sources of resistant bacteria and antibiotic residues, like pharmaceutical plants producing or manufacturing antibiotics.
In the first part of the project, we investigated the effects of waste effluent from a hospital (section 3.1) and a pharmaceutical plant manufacturing antibiotic products (section 3.2). The levels of susceptibility to six antimicrobial agents were determined in a total of 385 Acinetobacter isolates obtained from sites situated upstream (site A) and downstream (sites B and C) from the discharge points of the hospital (n=180) and the pharmaceutical plant (n=205).
In addition, a subset of 43 Acinetobacter isolates was characterised by biochemical tests and plasmid profiles to detect possible changes in the composition of the bacterial population associated with the discharge of waste effluent from the pharmaceutical plant (section 3.2). The methods used for sampling, bacterial isolation, antimicrobial susceptibility testing, phenotypic characterisation and plasmid profiling are described in chapter 2.
3.1 Effects of hospital waste effluent
Only minor differences were observed in the levels of antibiotic resistance between Acinetobacter isolates from sites situated upstream and downstream from the hospital discharge point (Table 3.1). Oxytetracycline resistance was detected only at sites B and C, and represented 37.2% and 12.5% of the total isolates from those sites, respectively. The occurrence of oxytetracycline-resistant isolates was significantly higher at site B compared with site A (P<0.01). However, at site C, 500 m beyond the discharge point, the levels of oxytetracycline resistance were significantly reduced compared with site B (P<0.01).
Surprisingly, the occurrence of chloramphenicol-resistant isolates observed at site B was significantly higher than that observed at site A (P<0.01). In relation to the other four antibiotics used for susceptibility testing (ampicillin, ciprofloxacin, gentamicin and sulfametoxazole), no significant differences were found between different sites in the occurrence of resistant isolates. Independent of the sampling site, most isolates were either susceptible to all antibiotics tested (51.7%) or resistant to only one compound (43.9%). Isolates resistant to more than two antibiotics were not detected at site A, and were rarely observed at sites B (1.3%) and C (4.7%)(Fig. 3.1).
Table 3.1.
Percentages of antibiotic resistance in 180 Acinetobacter isolates from the
sewers receiving waste effluent from a hospital.
Antibiotic |
% of resistant isolates |
||
Site A |
Site B |
Site C |
|
Amoxicillin |
2.6 |
2.6 |
3.1 |
Ciprofloxacin |
0 |
2.6 |
0 |
Chloramphenicol |
55.3a |
16.7 |
26.6 |
Gentamicin |
0 |
0 |
3.1 |
Oxytetracycline |
0 |
37.2 b |
12.5 c |
Sulfamethoxazole |
0 |
2.6 |
4.7 |
a
Statistically significant decrease in the numbers of resistant isolates occurring at site B compared with site A.a Statistically significant increase in the numbers of resistant isolates occurring at site B compared with site A.
c Statistically significant decrease in the numbers of resistant isolates occurring at site C compared with site B.
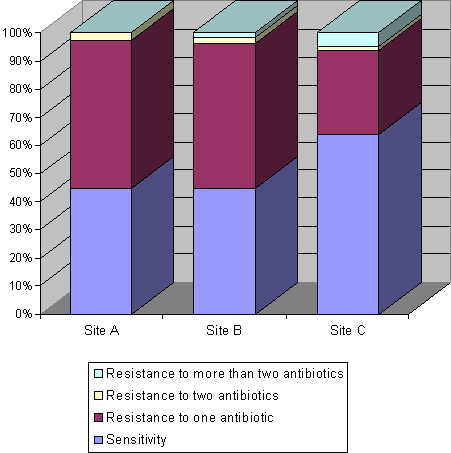
Figure 3.1.
Percentages of antibiotic sensitivity, resistance to one antibiotic and
multiple resistance in 180 Acinetobacter isolates from the sewers receiving waste effluent
from a hospital.
The reduction in the percentage of chloramphenicol resistance observed at downstream sites is difficult to explain. A possible explanation could be the presence of high numbers of chloramphenicol-sensitive strains in the hospital effluent, as this drug is no longer used in human medicine. Alternatively, substances present in the hospital effluent could affect the survival of chloramphenicol-resistant strains isolated upstream from the discharge point. Indeed, among the the 21 chloramphenicol-resistant isolates obtained from site A, 20 isolates were susceptible to the other five antibiotics used for susceptibility testing, and only a single isolate was resistant to amoxicillin.
The total numbers of culturable bacteria were markedly higher at the two sites B and C (106 to 107 CFU/ml) compared to site A (103 CFU/ml), indicating that the hospital effluent introduced high numbers of bacteria into the recipient sewers. Since the prevalence of Acinetobacter among total culturable bacteria was higher at sites B (23%) and C (9%) than at site A (<1%), it could be deduced that this organism was widely distributed in the hospital effluent. Indeed, it has recently been demonstrated that Acinetobacter is the most prevalent bacterial taxon (15% to 56%) among tetracycline-resistant heterotrophic bacteria in hospital waste effluent 60. These data confirm the suitability of our choice of Acinetobacter as a bacterial indicator for monitoring antibiotic resistance in hospital waste effluent.
The results of our study are in accordance with those of a previous study conducted at the same hospital by the former Danish Water Quality Institute (VKI) 61. Although different methods and target bacterial populations were used for monitoring of antibiotic resistance, both studies indicate a significant increase in the occurrence of tetracycline resistant bacteria associated with the discharge of waste effluent from the hospital. It is difficult to explain these results based on the data on national consumption of antibiotics in hospital care, as tetracyclines account for only a minor component of the antibiotics used in Danish hospitals in recent years 62. However, resistance to this antibiotic could also be co-selected by exposure to other substances, since tetracycline resistance genes are often located on plasmids together with other resistance genes.
3.2 Effects of pharmaceutical waste effluent
Acinetobacter isolates from the two sites situated downstream from the pharmaceutical plant demonstrated drastically increased levels of antibiotic resistance compared with isolates from the site situated upstream (Table 3.2). Significantly, higher percentages of Acinetobacter isolates resistant to amoxicillin, chloramphenicol, gentamicin, oxytetracycline and sulfamethoxazole were obtained from site B in comparison with site A (P<0.01). No statistically significant differences were detected between the two sites situated downstream from the sewage discharge point, indicating that the increase in the percentages of resistant isolates persisted at least 250 m from the discharge point.
Table 3.2.
Percentages of antibiotic resistance in 205 Acinetobacteer isolates from the
sewers receiving waste effluent from a pharmaceutical plant.
Antibiotic |
Antibiotic resistant isolates (%) |
||
Site A |
Site B |
Site C |
|
Amoxicillin |
10.5 |
26.4 a |
31.6 |
Ciprofloxacin |
1.3 |
0 |
0 |
Chloramphenicol |
25.0 |
51.4 a |
61.4 |
Gentamicin |
0 |
20.8 a |
12.3 |
Oxytetracycline |
5.3 |
48.6 a |
61.4 |
Sulfamethoxazole |
0 |
51.4 a |
57.9 |
a
Statistically significant increase in the numbers of resistant isolates occurring at site B compared with site A.High numbers of multiple-resistant Acinetobacter isolates were found at the sites situated downstream from the pharmaceutical plant (Fig. 3.2). Multiple resistance to three or more antibiotics was frequently observed among isolates from sites B (36.1%) and C (38.6%), whereas, this was not seen with isolates from site A. A statistically significant increase in the percentage of multiple-resistant isolates was shown for site B in comparison with site A (P<0.001), but only at the first three sampling times.
Resistance to sulfamethoxazole and oxytetracycline was often found associated among isolates from sites B (47.2%) and C (54.4%). This resistance pattern was observed in association with chloramphenicol resistance both among isolates from sites B (34.7%) and C (29.8%). Multiple resistance to all antibiotics used, except ciprofloxacin, was observed in 15.3% and 17.5% of the isolates from sites B and C, respectively. None of the above mentioned multiple resistance patterns were detected among isolates from site A.
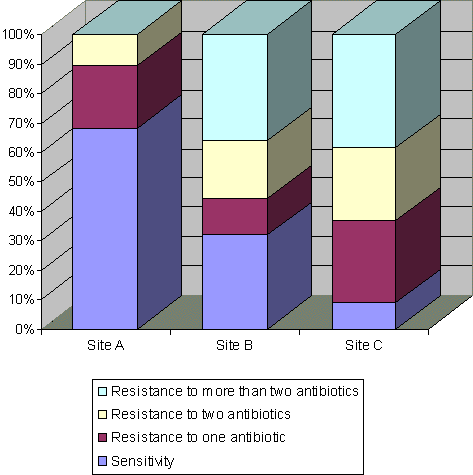
Figure 3.2.
Percentages of antibiotic sensitivity, resistance to one antibiotic and
multiple resistance in 205 Acinetobacter isolates from the sewers receiving waste effluent
from a pharmaceutical plant.
The observed increase in the prevalence of antibiotic resistance was associated with differences in the distribution of Acinetobacter species/strains between sites situated upstream and downstream from the pharmaceutical plant. Acinetobacter isolates from sites B and C showed different phenotypic patterns and plasmid profiles compared with isolates from site A. Seven different clones of Acinetobacter were found to be responsible for the high levels of multiple resistance observed at the two sites situated downstream from the pharmaceutical plant (Fig. 3.3).
According to phenotypic characterisation, the multiple-resistant strains isolated from the sewers receiving waste effluent from the pharmaceutical plant belonged to at least four different groups of Acinetobacter species: A. lwoffii/A. johnsonii (n=4), Acb complex (n=1), A. junii (n=1) and genomic species 16BJ (n=1). Some of the strains contained plasmids of similar size, but no single plasmid appeared to be present in all seven strains (Fig. 3.3). Consequently, the occurrence of multiple antibiotic resistance in the indigenous Acinetobacter population was not mediated by horizontal transfer of a unique plasmid structure.
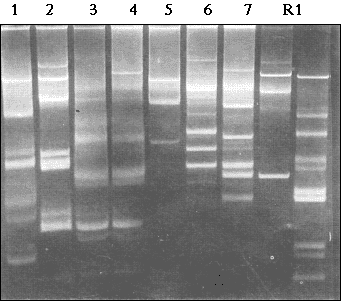
Figure 3.3.
Plasmid profiles of seven multiple-resistant Acinetobacter strains occurring
in the sewers situated downstream from the pharmaceutical plant (lanes 1-7). Lanes R1-R2,
reference strains E. coli 39R 861 and E. coli V517.
The combined use of antibiogram typing and plasmid profiling demonstrated that identical Acinetobacter strains were repeatedly isolated throughout the period of study. Isolates showing identical antibiotic resistance patterns, and identical or closely related plasmid profiles, were obtained from samples collected at different times or sites (Fig. 3.4). The clonal relationship of isolates showing identical plasmid profiles was further confirmed by Random Amplified Polymorphic DNA (RAPD) analysis, a DNA fingerprinting method for bacterial typing (data not shown). Based on these results, it could be concluded that multiple-resistant Acinetobacter clones, or presumptive clones, were established in the bacterial population of the sewers receiving waste effluent from the pharmaceutical plant.
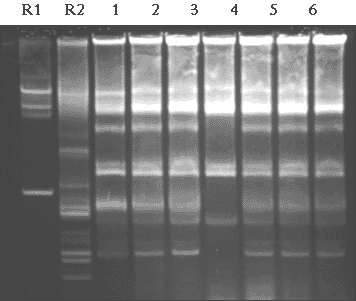
Figure 3.4.
Examples of identical or closely identical plasmid profiles in
Acinetobacter isolates obtained at different times or from different sites situated
downstream from the pharmaceutical plant (lanes 1-7). Lanes R1-R2, reference strains E.
coli 39R 861 and E. coli V517.
Previous studies on the effects of antibiotic resistance caused by the discharge of waste effluent from pharmaceutical plants, have been scarce. A brief report, previously conducted at the same pharmaceutical plant showed that the numbers of enterobacteria resistant to chloramphenicol, tetracyclines and aminoglycosides were higher at sites situated downstream from the effluent discharge point 63. A similar effect on antibiotic resistance was observed in sewers receiving waste effluent from another pharmaceutical plant producing fusidic acid 63. Therefore, according to the data currently available, it seems that waste effluent from pharmaceutical plants producing or manufacturing antibiotics enhances the occurrence of resistant bacteria in sewage.
The mechanisms by which waste effluent derived from antibiotic production or manufacture increases the numbers of resistant bacteria in sewage has not been investigated. At the pharmaceutical plant under study, products containing penicillins, tetracyclines, tylosin, and to a lesser extent aminoglycosides and sulfonamides, were manufactured during the period of sampling. At the end of each production cycle, the tanks where antibiotic agents had been produced were washed and the residual water, containing antibiotic residues and possibly resistant bacteria, was released directly into the sewage system. Consequently, the increased occurrence of resistant bacteria in the recipient sewers could be due to introduction of either antibiotic residues or resistant bacteria.
3.3 Conclusions
Waste effluent from the pharmaceutical plant was shown to have a higher impact on both single and multiple antibiotic resistance compared with waste effluent from the hospital, which had little effect on occurrence of resistant Acinetobacter in the recipient sewers. Therefore, waste effluent from pharmaceutical plants producing or manufacturing antibiotics could represent an important source for the occurrence of resistant bacteria in sewage.
This study indicates that human activities other than the indiscriminate use of antibiotics in human medicine, animal husbandry and agriculture, may disrupt the microbial balance in favour of resistant bacteria. Antibiotic resistance can develop not only in humans and in animals treated with antibiotics, but also in aquatic environments where antibiotics are present as residues derived from industrial production.
Further studies are necessary to assess the actual impact of antibiotic manufacturing on the spread of resistant bacteria in sewage. In particular, more pharmaceutical plants should be investigated, since various factors, such as type and size of production, could influence the occurrence of resistant bacteria and/or antibiotic residues in waste effluent derived from antibiotic production or manufacturing. Furthermore, studies should be implemented to investigate whether antibiotic residues originating from this source may negatively affect the biological treatment process at sewage treatment plants.